Mycoplasma cavipharyngis
(Hill, 1989)
Etymology
Gr. n. mukes – fungus, Gr. neut. n. plasma – anything formed, N.L. neut. n. Mycoplasma – fungus form; N.L. n. cavia – a guinea pig, N.L. n. pharynx – throat, N.L. gen. n. cavipharyngis – of the throat of a guinea pig
Taxonomy
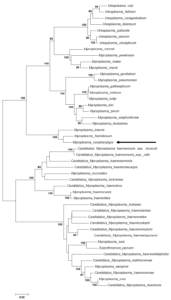
Mycoplasmatales – Mycoplasmataceae – Mycoplasma – Mycoplasma cavipharyngis (not part of a cluster), closely related to Mycoplasma fastidiosum (16S rRNA gene sequence similarity – 96.60%) (Fig. 1)
Type strain
117CT (guinea pig, UK, ≤1971), (Fig. 2, 16S rRNA gene sequence)
Genomes
no genome published (per 11/05/2024)
Cell morphology
coccoid – pleomorphic
Colony morphology
small and granular, fried egg morphology after prolonged incubation
Metabolism
fermentation of glucose; non-arginine-hydrolyzing, non-urea-hydrolyzing
Host
guinea pig
Habitat
pharynx, upper respiratory tract
Disease(s)
unknown, no disease reported
Pathogenicity
factors unknown
Epidemiology
unknown
Diagnosis
cultivation and species identification by MALDI-ToF MS or genetically
Fig. 1. Maximum likelihood tree showing the phylogenetic position of Mycoplasma cavipharyngis 117CT between the Pneumoniae/Ureaplasma and Hemoplasma cluster of Mycoplasmataceae based on 16S rRNA gene sequences. Numbers at nodes represent bootstrap confidence values (1000 replications). Only values > 80% are shown. Bar, number of substitutions per nucleotide position. Credits: Joachim Spergser (Vetmeduni Vienna)