Mycoplasma agalactiae
(Wroblewski, 1931; Freundt, 1955)
Etymology
Gr. n. mukes – fungus, Gr. neut. n. plasma – anything formed, N.L. neut. n. Mycoplasma – fungus form; Gr. n. agalactia – want of milk, N.L. gen. n. agalactiae – of agalactia
Taxonomy
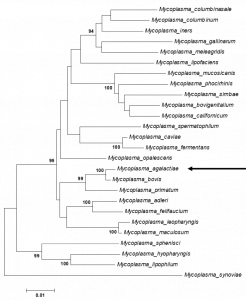
Mycoplasmatales – Mycoplasmataceae – Mycoplasma – Mycoplasma agalactiae (Bovis cluster), closely related to Mycoplasma bovis (16S rRNA gene sequence similarity – 99.45%) (Fig. 1)
Type strain
PG2T (goat, Spain, 1959), (Fig. 2, 16S rRNA gene sequence)
Genomes
10 completed (PG2T – Spain; 5623 – Spain; JF4428 (clone of PG2T) – Spain; GrTh01 – Greece; GM139 – USA; HOWD44, GA572, GA568, BAH356 – Mongolia; 2463 – Germany); 11 draft genomes (NCBI Genome deposits per 11/05/2024)
Cell morphology
spherical – coccoid
Colony morphology
fried egg morphology (Fig. 3)
Metabolism
oxidation of organic acids (pyruvate, lactate); non-fermentative, non-arginine-hydrolyzing, non-urea-hydrolyzing
Host
sheep and goats, occasionally reported in wild ungulates
Habitat
mainly udder, rarely upper respiratory tract
Disease(s)
major cause of Contagious Agalactia (CA) presenting mastitis with complete loss of milk production; arthritis, keratoconjunctivitis, pneumonia and septicemia in young animals
Pathogenicity
known pathogenicity factors - a family of phase- and size-variable membrane surface lipoproteins (Vpma) with role in adhesion, cell invasion and immune evasion, P40 adhesin, P48 macrophage stimulatory protein, Opp involved in biofilm formation and adaptation to specific environments
Epidemiology
reported to occur in Mediterranean countries, the Balkans, India, USA, and South America; transmission by milk (at suckling, by milkers’ hands, milking machine, bedding) or via aerosol; commonly introduced into herds by latently infected carrier animals
Diagnosis
cultivation and species identification by MALDI-ToF MS, serology or genetically; PCR; detection of antibodies using immunoassays for herd testing
Fig. 1. Maximum likelihood tree showing the phylogenetic position of Mycoplasma agalactiae PG2T within the Bovis cluster of Mycoplasmataceae based on 16S rRNA gene sequences. The sequence of Mycoplasma synoviae WVU 1853T was used as out-group (Synoviae cluster). Numbers at nodes represent bootstrap confidence values (1000 replications). Only values > 80% are shown. Bar, number of substitutions per nucleotide position. Credits: Joachim Spergser (Vetmeduni Vienna)
CTGGCTGTGTGCCTAATACATGCATGTCGAGCGATGATAGCAATATCATAGCGGCGAATGGGTGAGTAACACGTACTCAACGTACCTTTTAGATTGGGATAGCGGATGGAAACATCCGATAATACAGAATACTTATTATTTTTGCATGAAAGTAATATAAAAGGAAGCGTTTGCTTCGCTAGAAGATCGGAGTGCGCAACATTAGCTAGTTGGTGAGGTAACGGCCCACCAAGGCGATGATGTTTAGCGGGGTTGAGAGATTGATCCGCCACACTGGGACTGAGATACGGCCCAGACTCCTACGGGAGGCAGCAGTAGGGAATATTCCACAATGGACGAAAGTCTGATGGAGCGACACAGCGTGCAGGATGAAGGCCCTATGGGTTGTAAACTGCTGTGGTTAGGGAAGAAAAAGTAGCGTAGGAAATGACGCTACCTTGACGGTACCTGATTAGAAAGCAACGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGTTGCAAGCGTTATCCGAAATTATTGGGCGTAAAGCGTCTGTAGGTTGTTTGTTAAGTCTGGCGTTAAATTTTGGGGCTCAACCCCAAAACGCGTTGGATACTGGCAGACTAGAGTTATGTAGAGGTTAGCGGAATTCCTTGTGAAGCGGTGAAATGCGTAGATATAAGGAAGAACATCAATATGGCGAAGGCAGCTAACTGGGCATACACTGACACTGAGAGACGAAAGCGTGGGGAGCAAACAGGATTAGATACCCTGGTAGTCCACGCCCTAAACGATGATCATTAGTTGATGGGGAACTCATCGACGCAGCTAACGCATTAAATGATCCGCCTGAGTAGTACGTTCGCAAGAATAAAACTTAAAGGAATTGACGGGGATCCGCACAAGCGGTGGAGCATGTGGTTTAATTTGAAGATACGCGTAGAACCTTACCCACTCTTGACATCTTCTGCAAAGCTATGGAGACATAGTGGAGGTTAACAGAATGACAGATGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTCGGTTAAGTCCTACAACGAGCGCAACCCTTATCCTTAGTTACTACCATTTAGTTGAGCACTCTAAGGAGACTGCCCGAGTAATTGGGAGGAAGGTGGGGACGACGTCAAATCATCATGCCTCTTACGAGTGGGGCTACACACGTGCTACAATGGACGGTACAAAGAGAAGCGAAGTGGTGACATGGAGCAAACCTCAAAAAACCGTTCTCAGTTCGGATTGAAGTCTGCAACTCGACTTCATGAAGTCGGAATCGCTAGTAATCGTAGATCAGCTACGCTACGGTGAATACGTTCTCGGGTCTTGTACACACCGCCCGTCAAACCATGGGAGCTGGTAATGCCCGAAGTCGGTTTATTAAGAAACTGCCTAAGGCAGGACTGGTGACTGGGGTTAAGTCGTAACAAGGT
Fig. 2. 16S rRNA gene sequence of Mycoplasma agalactiae PG2T (Accession number: NR_118811)Fig. 3. Colonies of Mycoplasma agalactiae PG2T on modified Hayflick’s agar after 4 days of incubation exhibiting characteristic fried egg morphology. Bar, 1 mm. Credits: Joachim Spergser (Vetmeduni Vienna)