Anaeroplasma bactoclasticum
(Robinson & Hungate, 1973; Robinson & Allison, 1975)
Etymology
Gr. pref. a – not, without, Gr. n. aer – air, Gr. neut. n. plasma – anything formed, N.L. neut. n. Anaeroplasma – a form not living with air; N.L. dim. n. bact- – part of N.L. n. bacterium – a small rod, N.L. adj. clasticus – breaking, N.L. neut. adj. bactoclasticum –bacteriolytic (referring to the organisms’ ability to digest bacterial cells)
Taxonomy
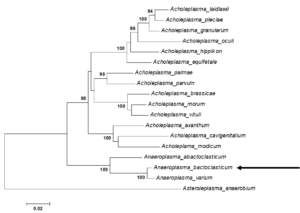
Anaeroplasmatales – Anaeroplasmataceae – Anaeroplasma – Anaeroplasma bactoclasticum, closely related to Anaeroplasma varium (16S rRNA gene sequence similarity – 97.77%) (Fig. 1)
Type strain
JRT (not available, initially isolated from bovine rumen, USA, ≤1972), (Fig. 2, 16S rRNA gene sequence)
Genomes
4 draft genomes (material from JRT – USA, 3 from metagenomics – USA) (NCBI Genome deposit per 11/05/2024)
Cell morphology
spherical – coccoid
Colony morphology
fried egg morphology
Metabolism
fermentation of glucose; non-arginine-hydrolyzing, non-urea-hydrolyzing
Host
ruminants
Habitat
rumen
Disease(s)
natural inhabitant of the rumen
Pathogenicity
apathogenic
Epidemiology
unknown
Diagnosis
anaerobic cultivation and genetic species identification
Fig. 1. Maximum likelihood tree showing the phylogenetic position of Anaeroplasma bactoclasticum JRT close to genus Acholeplasma based on 16S rRNA gene sequences. Numbers at nodes represent bootstrap confidence values (1000 replications). Only values > 80% are shown. Bar, number of substitutions per nucleotide position. Credits: Joachim Spergser (Vetmeduni Vienna)